sessionInfo()
# R version 4.3.1 (2023-06-16 ucrt)
# Platform: x86_64-w64-mingw32/x64 (64-bit)
# Running under: Windows 11 x64 (build 26100)
#
# Matrix products: default
#
#
# locale:
# [1] LC_COLLATE=Portuguese_Brazil.utf8 LC_CTYPE=Portuguese_Brazil.utf8
# [3] LC_MONETARY=Portuguese_Brazil.utf8 LC_NUMERIC=C
# [5] LC_TIME=Portuguese_Brazil.utf8
#
# time zone: America/Sao_Paulo
# tzcode source: internal
#
# attached base packages:
# [1] stats4 stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] biscale_1.0.0 cowplot_1.1.3 rmapshaper_0.5.0 sf_1.0-14
# [5] rnaturalearth_1.0.1 DHARMa_0.4.7 bbmle_1.0.25.1 performance_0.15.0
# [9] glmmTMB_1.1.11 countrycode_1.6.1 tidyr_1.3.0 patchwork_1.2.0
# [13] ggplot2_3.5.0.9000 scales_1.4.0 dplyr_1.1.3 tibble_3.2.1
# [17] ggeffects_2.3.0 here_1.0.1 readr_2.1.4
#
# loaded via a namespace (and not attached):
# [1] RColorBrewer_1.1-3 rstudioapi_0.15.0 jsonlite_1.9.0 datawizard_1.1.0
# [5] magrittr_2.0.3 farver_2.1.1 nloptr_2.0.3 ragg_1.2.6
# [9] vctrs_0.6.4 minqa_1.2.6 terra_1.7-55 forcats_1.0.0
# [13] htmltools_0.5.7 itertools_0.1-3 clustMixType_0.4-2 haven_2.5.3
# [17] curl_5.1.0 betapart_1.6 KernSmooth_2.23-21 plyr_1.8.9
# [21] TMB_1.9.17 igraph_2.0.2 mime_0.12 lifecycle_1.0.4
# [25] minpack.lm_1.2-4 iterators_1.0.14 pkgconfig_2.0.3 sjlabelled_1.2.0
# [29] phyloregion_1.0.8 gap_1.6 Matrix_1.6-1.1 R6_2.5.1
# [33] fastmap_1.1.1 snakecase_0.11.1 rbibutils_2.3 shiny_1.8.0
# [37] magic_1.6-1 digest_0.6.33 numDeriv_2016.8-1.1 colorspace_2.1-0
# [41] rprojroot_2.0.4 textshaping_0.3.7 qgam_2.0.0 vegan_2.7-1
# [45] labeling_0.4.3 fansi_1.0.5 httr_1.4.7 abind_1.4-5
# [49] mgcv_1.9-3 compiler_4.3.1 proxy_0.4-27 bit64_4.0.5
# [53] withr_2.5.2 doParallel_1.0.17 DBI_1.1.3 MASS_7.3-60
# [57] classInt_0.4-10 permute_0.9-8 units_0.8-4 tools_4.3.1
# [61] ape_5.8-1 httpuv_1.6.13 glue_1.6.2 quadprog_1.5-8
# [65] rcdd_1.6 nlme_3.1-162 promises_1.2.1 grid_4.3.1
# [69] cluster_2.1.4 generics_0.1.3 snow_0.4-4 predicts_0.1-19
# [73] gtable_0.3.4 tzdb_0.4.0 class_7.3-22 hms_1.1.3
# [77] sp_2.1-3 utf8_1.2.4 foreach_1.5.2 pillar_1.9.0
# [81] vroom_1.6.4 later_1.3.1 splines_4.3.1 lattice_0.21-8
# [85] bit_4.0.5 tidyselect_1.2.0 knitr_1.45 reformulas_0.4.1
# [89] V8_6.0.4 xfun_0.41 smoothr_1.1.0 geojsonsf_2.0.3
# [93] boot_1.3-28.1 codetools_0.2-19 maptpx_1.9-7 cli_3.6.1
# [97] xtable_1.8-4 geometry_0.5.2 systemfonts_1.2.1 Rdpack_2.6.4
# [101] Rcpp_1.0.11 doSNOW_1.0.20 bdsmatrix_1.3-7 parallel_4.3.1
# [105] picante_1.8.2 ellipsis_0.3.2 gap.datasets_0.0.6 lme4_1.1-35.1
# [109] phangorn_2.12.1 mvtnorm_1.2-3 slam_0.1-50 e1071_1.7-13
# [113] insight_1.3.1 purrr_1.0.2 crayon_1.5.2 combinat_0.0-8
# [117] rlang_1.1.2 fastmatch_1.1-6The hidden biodiversity knowledge split in biological collections
This manuscript is submitted as a preprint here
General overview
This repository contains the data and code used in the analysis of the manuscript entitled “The hidden biodiversity knowledge split in biological collections”.
In this study we characterized different aspects of spatial and temporal patterns of fish Name Bearing Types (NBT) among countries and world regions. The characteristics comprises the number of total NBT, the NBT flowing among different world regions, the characteristics of regions and countries regarding the source of NBT in their biological collection, the level of underepresentation of native species and the level of overepresentation of non-native species for each country.
We discuss how the fundamental knowledge in fish species is distributed and its implications for science development and knowledge sharing.
Repository structure
data
This folder stores raw and processed data used to perform all the analysis presented in this study
raw
flow_period_region_country.csva data frame in the long format containing the flowing of NBT per regions per per time (50-year time frame). Variables:periodnumeric variable representing 50-year time intervalsregion_typecharacter representing the name of the World Bank region of the country where the NBT was sourcedcountry_typecharacter. A three letter code (alpha-3 ISO3166) representing the country of the museum where the NBT was sourcedregion_museumcharacter. Name of the World Bank region of the country where the NBT is housedcountry_museumcharacter. A three letter code (alpha-3 ISO3166) representing the country of the museum where the NBT is housednnumeric. The number of NBT flowing from one country to another
spp_native_distribution.csvdata frame in the long format containing the native composition at the country level. Variables:speciescharacter. The name of a species in the format genus_epithet according to the Catalog of Fishes (including synonym names)country_distributioncharacter. Three letter code (alpha-3 ISO3166) indicating the name of the country where a species is native toregion_distributioncharacter. The name of the region acording with World Bank where a species is native to
spp_type_distribution.csvdata frame in the long format containing the composition of NBT by country. Variables:speciescharacter. The name of a species in the format genus_epithet according to the Catalog of Fishes (including synonym names)country_distributioncharacter. Three letter code (alpha-3 ISO3166) indicating the name of the country where a species is housedregion_distributioncharacter. The name of the region acording with World Bank where a species is housed
bio-dem_data.csvdata frame with data downloaded from Bio-Dem containing information on biological and social information at the country level. Variables:countrycharacter. A three letter code (alpha-3 ISO3166) representing a countryrecordsnumeric. Total number of species occurrence records from Global Biodiverity Facility (GBIF)records_per_areanumeric. Records per area from gbifyearsSinceIndependencenumeric. Years since independence for each countrye_migdppcnumeric. GDP per capta
museum_data.csvdata frame with museums’ acronyms and the world region of each. Variables:code_museumcharacter. Three letter code of the museumcountry_museumcharacter. A three letter code (alpha-3 ISO3166) representing a countryregion_museumcharacter. The name of the region acording with World Bank
processed
flow_region.csva data frame containing flowing of NBT among world regions and the total number of NBT derived from the source regionregion_typecharacter. The region, according to World bank classification, where the type was collectedregion_museumcharacter. The region, according to World bank classification, where the type is housednnumeric. The number of types that flowed fromregion_typetoregion_museumtotal_region_typenumeric. The total number of name bearers sampled in each region
flow_period_region.csva data frame with the number of NBT between the world regions per 50-year time frame and the total number of NBT in each time frame for each world regionperiodnumeric. The year in which the name bearer was discoveredregion_typecharacter. The region, according to World bank classification, where the type was collectedregion_museumcharacter. The region, according to World bank classification, where the type is housednnumeric. The number of name bearers that flowed fromregion_typetoregion_museumtotal_region_typenumeric. The total number of name bearers sampled in each region in each time period
flow_period_region_prop.csva data frame with the number of NBT, the Domestic Contribution and Domestic Retention between the world regions in a 50-year time frameperiodnumeric. The year in which the name bearer was discoveredregion_typecharacter. The region, according to World bank classification, where the type was collectedregion_museumcharacter. The region, according to World bank classification, where the type is housednnumeric. The number of name bearers that flowed fromregion_typetoregion_museumtotal_period_region_typenumeric. The number of name bearers samples in a region in a time periodtotal_period_region_museumnumeric. The number of name bearers housed in biological collections in a region in a time periodtotal_periodnumeric. The total number of name bearers described in a periodprop_DCnumeric. The proportion of name bearers in biological collection of a region sampled within the region in a time period. This variable is not used in the final version of the study.prop_DRnumeric. The proportion of all name bearers sampled that were retained in a region in a time period. This variable is not used in the final version of the study.
flow_region_prop.csvdata with the total number of species flowing between world regions, Domestic Contribution and Domestic Retentionregion_typecharacter. The region, according to World bank classification, where the type was collectedregion_museumcharacter. The region, according to World bank classification, where the type is housedtotal_region_typenumeric. The total number of name bearers sampled in a regiontotal_region_museumnumeric. The total number of name bearers housed in biological collections in a regionprop_DCnumeric. The proportion of name bearers in biological collection of a region sampled within the region. This variable is not used in the final version of the studyprop_DRnumeric. The proportion of all name bearers sampled that were retained in a region. This variable is not used in the final version of the study
flow_country.csvdata frame with flowing information of NBT among countriescountry_typecharacter. A three letter code (alpha-3 ISO3166) representing a country where the name bearer was sampledcountry_museumcharacter. A three letter code (alpha-3 ISO3166) representing a country where the name bearer was housednnumeric. The number of name bearers that flowed fromcountry_typetocountry_museumtotal_country_typenumeric. The number of name bearers sampled in a country
df_country_native.csvdata frame with the number of native species at the country levelcountry_distributioncharacter. A three letter code (alpha-3 ISO3166) representing a countryregion_distributioncharacter. The name of a region according to World Banknative.richnessnumeric. The number of native species in a country according to Catalog of Fishes
df_country_type.csvdata frame with the number of NBT at the country levelcountry_museumcharacter. A three letter code (alpha-3 ISO3166) representing a countryregion_museumcharacter. The name of a region according to World Banktype_regionnumeric. The number of name bearers in biological collections in countries
df_endemic_beta.csvdata frame with values of endemic deficit and non-endemic representation at the country level using only species with restricted occurrences (only one occurrence per country) at the country levelnative.betanumeric. The endemic deficit calculated as the number of endemic species outside of the country of origintype.betanumeric. The number of non-endemic name bearers
df_all_beta.csvdata frame with values of endemic deficit and non-endemic representation at the country level. This is used in the analysis of Supplementary material, specifically to generate Figure S2.countriescharacter. A three letter code (alpha-3 ISO3166) representing a countrynative.betanumeric. The endemic deficit calculated as the number of endemic species outside of the country of origintype.betanumeric. The number of non-endemic name bearers
R
The letters D, A and V represents scripts for, respectively, data processing (D), data analysis (A) and results visualization (V). The script sequence to reproduce the workflow is indicated by the numbers at the beginning of the name of the script file.
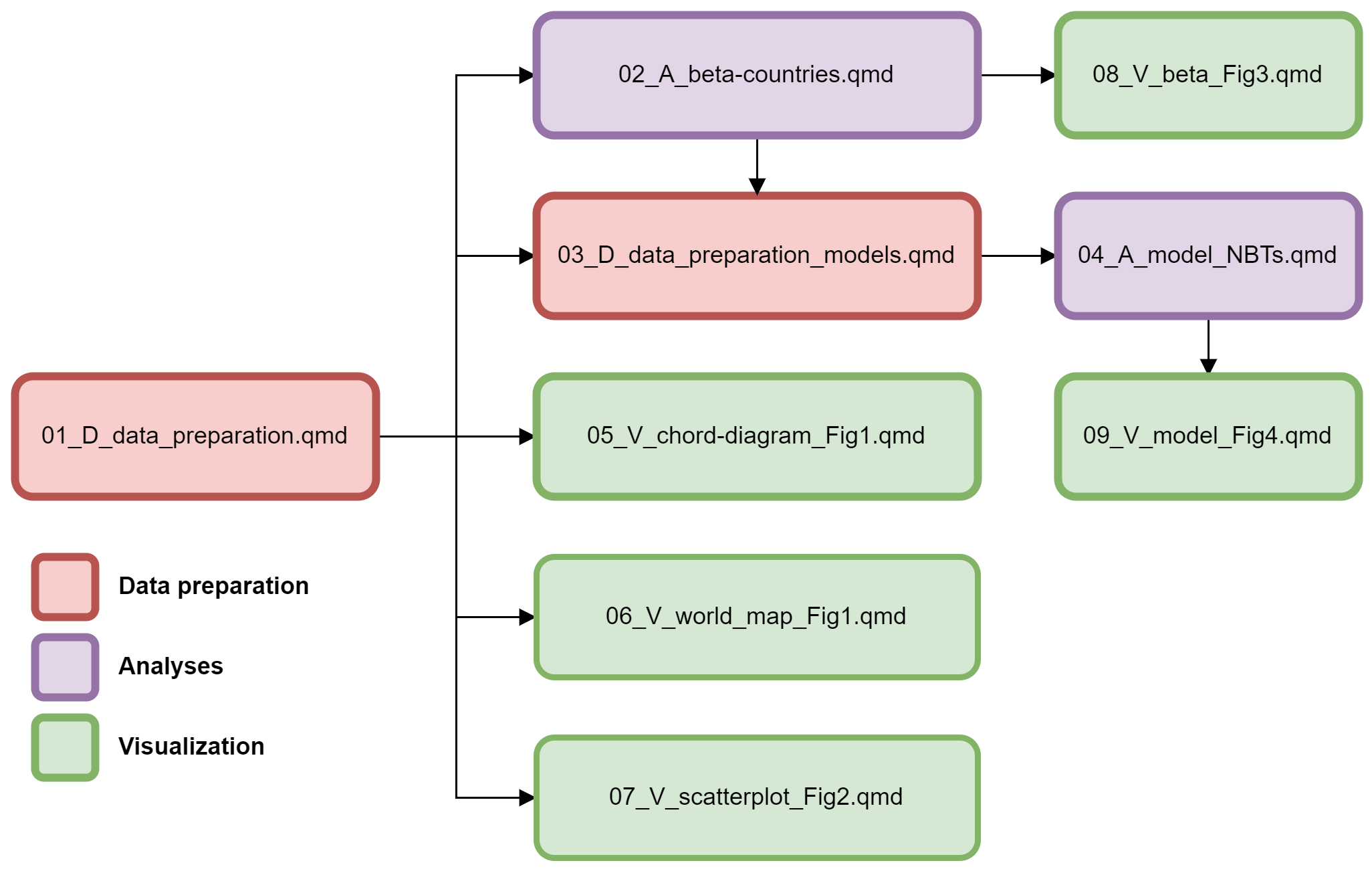
01_D_data_preparation.qmdinitial data preparation02_A_beta-endemics-countries.qmdThis script is used to calculatenon endemic name bearersandendemic deficitthat will be used in the script07_V_beta_endemics_Fig2.qmd03_D_data_preparation_models.qmdscript used to build data frames that will be used in statistical models (04_A_model_NBTs.qmd)04_A_model_NBTs.qmdstatistical models for the total number of NBT, endemic deficit and non endemic name bearers05_V_chord_diagram_Fig1.qmdcode used to produce circular flow diagram. This is the Figure 1 of the study06_V_world_map_Fig1.qmdcode used to produce the world map in the Figure 1 of the main text07_V_beta_endemics_Fig2.qmd code used to build Figure 2 of the main text
08_V_model_Fig3.qmdcode used to build the Figure 3 of the main text. This is the representation of the results of the models present in the script 04_A_model_NBTs.qmd09_Supplementary_analysis.qmdcode to produce all the tables and figures presented in the Supplementary material of this study
functions
function_beta_types_success_fail.Rfunction used to calculate endemic deficit and non endemic name bearers.function_scale_back.Rfunction used to transform back normalized variables
Summary statistics
011_Summary_stats.qmdcode needed to reproduce summary statistics reported in the Results section of the main text of the study “The hidden biodiversity knowledge split in biological collections”
output
Figures
In this folder you will find all figures used in the main text and supplementary material of this study
Fig1_flow_circle_plot.pngFigure with circular plots showing the flux of NBT among regions of the world in a 50-year time window
Fig2_turnover_metrics_endemics.pngCartogram with 3 maps showing the level of endemic deficit, non endemic name bearers and the combination of both metrics biscale map. This is the Figure 2 in the main text
Fig3_models.png Figure showing the predictions of the number of name bearers, endemic deficit and non endemic name bearers for different predictors. Corresponds to Figure 3 This is derived from the statistical models scripts
Supp-material
This folder contains the figures in the Supplementary material
FigS1_native_richness.pngWorld map with countries colored according to the number of native species richness according to the Catalog of Fishes. This corresponds to Figure S1 in Supplementary materialFigS3_turnover_metrics.pngCartogram with 3 maps showing the level of native turnover, NBT turnover and the combination of both metrics in a combined map. This corresponds to Figure S2 in Supplementary material
Packages
| Package | Version | Documentation |
|---|---|---|
| bbmle | 1.0.25.1 | bbmle |
| betapart | 1.6 | betapart |
| biscale | 1.0.0 | biscale |
| circlize | 0.4.15 | circlize |
| countrycode | 1.6.0 | countrycode |
| cowplot | 1.1.1 | cowplot |
| DHARMa | 0.4.6 | DHARMa |
| dplyr | 1.1.4 | dplyr |
| ggarrow | 0.0.0.9000 | ggarrow |
| ggplot2 | 3.5.0 | ggplot2 |
| glmmTMB | 1.1.8 | glmmTMB |
| glue | 1.6.2 | glue |
| here | 1.0.1 | here |
| patchwork | 1.2.0.9000 | patchwork |
| performance | 0.12.1 | performance |
| phyloregion | 1.0.8 | phyloregion |
| readr | 2.1.4 | readr |
| rmapshaper | 0.5.0 | rmapshaper |
| rnaturalearth | 0.3.4 | rnaturalearth |
| scales | 1.4 | scales |
| sf | 1.0-14 | sf |
| tidyr | 1.3.1 | tidyr |
Contact
Gabriel Nakamura and Bruno Mioto
If you have any suggestion or commentary, please open an issue