library(circlize) #chord diagrams
library(dplyr) #data manipulation
library(glue) #string interpolation05 - Producing chord diagram (Figure 1)
05_V_chord_diagram_Fig1.qmd
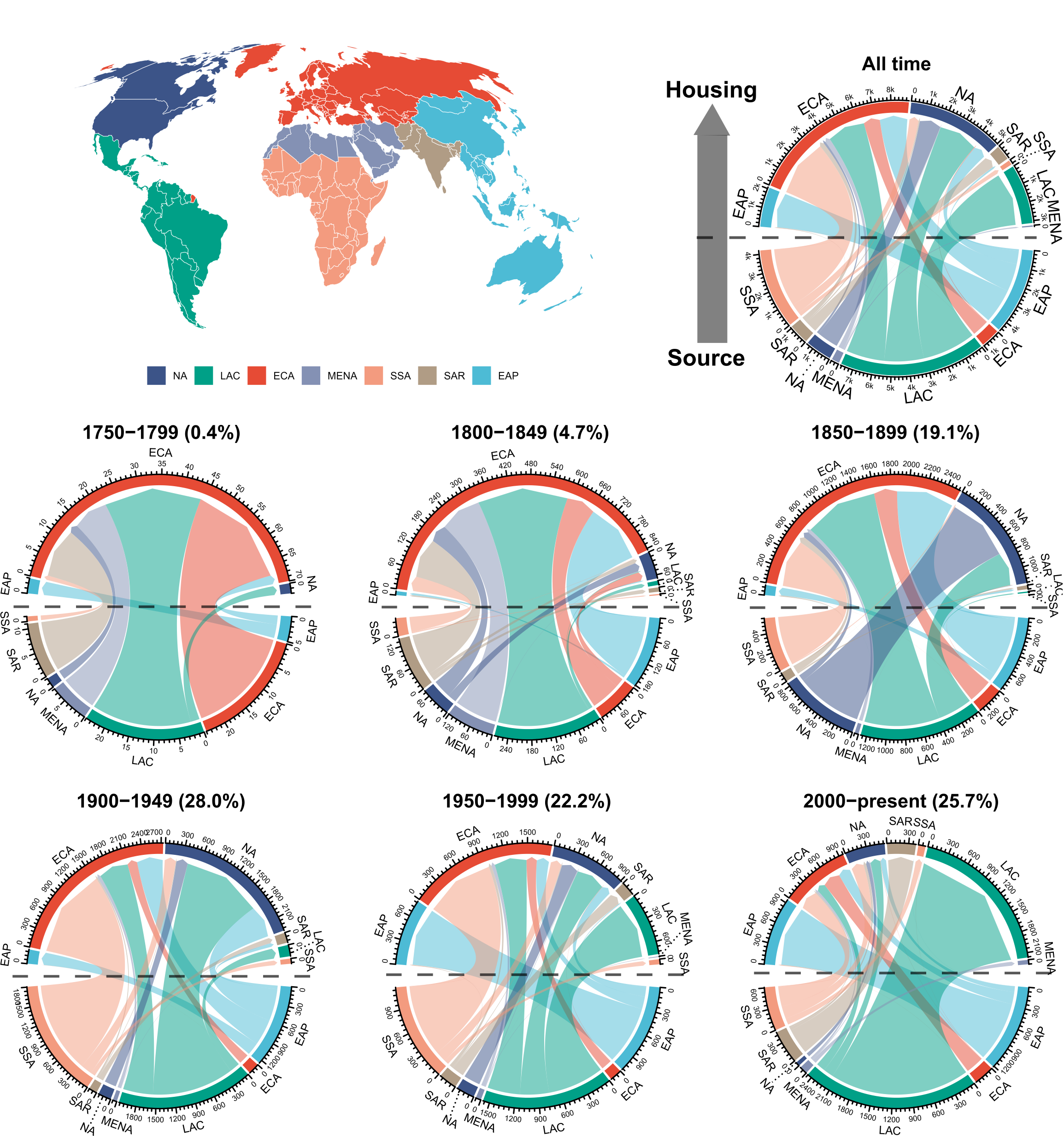
Here we produce the chord diagram (Figure 1 in the manuscript)
Data used in this script
# Data from 01_C_data_preparation.qmd
flow_period_region <- readr::read_csv(here::here("data", "processed", "flow_period_region.csv"))
flow_region <- readr::read_csv(here::here("data", "processed", "flow_region.csv")) Mapping name bearers flow among regions per time interval of 50 years
# use region abbreviation
flow_period_region2 <- flow_period_region |>
dplyr::mutate(
dplyr::across(
c(region_type, region_museum),
~dplyr::case_when(
. == "Europe & Central Asia" ~ "ECA",
. == "East Asia & Pacific" ~ "EAP",
. == "North America" ~ "NA",
. == "South Asia" ~ "SAR",
. == "Latin America & Caribbean" ~ "LAC",
. == "Sub-Saharan Africa" ~ "SSA",
. == "Middle East & North Africa" ~ "MENA"
)))
flow_region2 <- flow_region |>
dplyr::mutate(
dplyr::across(
c(region_type, region_museum),
~dplyr::case_when(
. == "Europe & Central Asia" ~ "ECA",
. == "East Asia & Pacific" ~ "EAP",
. == "North America" ~ "NA",
. == "South Asia" ~ "SAR",
. == "Latin America & Caribbean" ~ "LAC",
. == "Sub-Saharan Africa" ~ "SSA",
. == "Middle East & North Africa" ~ "MENA"
)))
# create dataset for each region
regions_1750 <- flow_period_region2 |>
dplyr::filter(period == 1750) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(
value != 0
)
regions_1800 <- flow_period_region2 |>
dplyr::filter(period == 1800) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(
value != 0
)
regions_1850 <- flow_period_region2 |>
dplyr::filter(period == 1850) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(
value != 0
)
regions_1900 <- flow_period_region2 |>
dplyr::filter(period == 1900) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(
value != 0
)
regions_1950 <- flow_period_region2 |>
dplyr::filter(period == 1950) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(
value != 0
)
regions_2000 <- flow_period_region2 |>
dplyr::filter(period == 2000) |>
dplyr::select(-c(period, total_period_region_type)) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(value != 0)
regions_alltime <- flow_region2 |>
dplyr::select(-total_region_type) |>
dplyr::mutate(
region_type = glue::glue(" {region_type}"),
region_museum = glue::glue("{region_museum} ")) |>
dplyr::rename(
from = region_type,
to = region_museum,
value = n) |>
dplyr::filter(value != 0)
#define colors
colors <- c(
" ECA" = "#f34d4d",
" EAP" = "#4da8a6",
" NA" = "#416295",
" SAR" = "#a6a7a6",
" LAC" = "#4dae7d",
" SSA" = "#E0A0A7",
" MENA" = "#9D7AAB",
"ECA " = "#f34d4d",
"EAP " = "#4da8a6",
"NA " = "#416295",
"SAR " = "#a6a7a6",
"LAC " = "#4dae7d",
"SSA " = "#E0A0A7",
"MENA " = "#9D7AAB"
)Producing the figures
#start pdf
pdf(here::here("output", "figures", "Fig1.pdf"), width = 8, height = 8)
#define layout
# The layout will follow this order
# 1 4 7
# 2 5 8
# 3 6 9
layout(matrix(1:9, 3, 3))
#1 - blank space for map
plot(0, type='n', axes=FALSE, ann=FALSE)
#2 - 1750-1799
chordDiagram(regions_1750,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("1750-1799")
#3 - 1900-1949
chordDiagram(regions_1900,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("1900-1949")
#4 - blank space for map
plot(0, type='n', axes=FALSE, ann=FALSE)
#5 - 1800-1849
chordDiagram(regions_1800,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("1800-1849")
#6 - 1950-1999
chordDiagram(regions_1950,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("1950-1999")
#7 - All time
chordDiagram(regions_alltime,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
annotationTrack = "grid",
reduce = 0.000000000000001,
preAllocateTracks = list(track.height = 0.1,
track.margin = c(0,0)),
annotationTrackHeight = mm_h(c(2, 2)),
)
circos.track(track.index = 1, panel.fun = function(x, y) {
if(abs(CELL_META$cell.start.degree - CELL_META$cell.end.degree) > 0) {
sn = CELL_META$sector.index
i_state = as.numeric(gsub("(C|R)_", "", sn))
circos.text(CELL_META$xcenter, 1, CELL_META$sector.index,
facing = "inside", niceFacing = TRUE, adj = c(0.5,0))
xlim = CELL_META$xlim
breaks = seq(0, 10000, by = 1000)
circos.axis(
major.at = breaks,
labels = ifelse(breaks >= 1000, paste0(breaks/1000, "k"), breaks),
labels.cex = 0.5,
h = "bottom"
)
}
}, bg.border = NA)
title("All time")
#8 - 1850-1899
chordDiagram(regions_1850,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("1850-1899")
#9 - 2000-present
chordDiagram(regions_2000,
grid.col = colors,
directional = 1,
direction.type = c("arrows"),
link.arr.type = "big.arrow",
reduce = 0.000000000000001,
)
title("2000-present")
#Finish
dev.off()png
2 circos.clear()Plotting the final figure
Note
The manuscript version of this Figure is edited with Inkscape software
This is the final figure that correspond to the Figure 1 of the manuscript