#plot
library(ggplot2)
library(patchwork)
library(cowplot)
#map
library(rnaturalearth)
library(rmapshaper)
library(sf)
library(biscale)07 - World map with countries represented by its endemic deficit for non-endemic representation (Figure 2)
07_V_beta_endemics_Fig2.qmd
Plotting endemic deficit and non-endemic representation for each country
Loading packages
Data used
# Data from 02_D_beta-countries.qmd
df_endemic_beta <- readr::read_csv(here::here("data", "processed", "df_endemic_beta.csv"))Joining metric information with geographical data
countries <- rnaturalearth::ne_countries(returnclass = "sf")
sf_countries <-
sf::st_as_sf(countries) |>
dplyr::filter(admin != "Antarctica") |>
sf::st_transform(crs = "+proj=moll +x_0=0 +y_0=0 +lat_0=0 +lon_0=0") |>
dplyr::select(iso_a3_eh)
df_endemic_beta_sf <-
sf_countries |>
dplyr::full_join(df_endemic_beta, by = c(iso_a3_eh = "countries"))First processing spatial data to convert NA values into 0
df_endemic_beta_sf2 <-
df_endemic_beta_sf |>
sf::st_as_sf() |>
rmapshaper::ms_filter_islands(min_area = 12391399903) |>
dplyr::mutate(
type.beta = ifelse(is.na(type.beta),
0,
type.beta),
native.beta = ifelse(is.na(native.beta),
0,
native.beta))Create palettes
palette_blue <- colorRampPalette(c("#d3d3d3", "#accaca", "#81c1c1", "#52b6b6"))
palette_pink <- colorRampPalette(c("#d3d3d3", "#c5acc2", "#bb84b1", "#ac5a9c"))Plotting maps
map_native_endemic_beta <-
ggplot() +
geom_sf(data = df_endemic_beta_sf2,
aes(geometry = geometry,
fill = native.beta),
color = "white",
size = 0.1, na.rm = T) +
scale_fill_gradientn(
colors = palette_pink(10),
na.value = "#d3d3d3",
limits = c(0,1)
)+
guides(fill = guide_colorbar(
barheight = unit(0.1, units = "in"),
barwidth = unit(4, units = "in"),
ticks.colour = "grey20",
title.position="top",
title.hjust = 0.5
)) +
labs(
fill = "Endemic deficit"
)+
theme_classic()+
theme(
legend.position = "bottom",
legend.margin = margin(-10,0,0,0,"pt"),
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.line = element_blank()
)
map_type_endemic_beta <-
ggplot() +
geom_sf(data = df_endemic_beta_sf2,
aes(geometry = geometry,
fill = type.beta),
color = "white",
size = 0.1, na.rm = T) +
scale_fill_gradientn(
colors = palette_blue(10),
na.value = "#d3d3d3",
limits = c(0,1)
)+
guides(fill = guide_colorbar(
barheight = unit(0.1, units = "in"),
barwidth = unit(4, units = "in"),
ticks.colour = "grey20",
title.position="top",
title.hjust = 0.5
)) +
labs(
fill = "Non-endemic name bearers"
)+
theme_classic()+
theme(
legend.position = "bottom",
legend.margin = margin(-10,0,0,0,"pt"),
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.line = element_blank()
) Plotting the two quantities in a bivariate map
sf_bivar_types_endemic <-
bi_class(df_endemic_beta_sf2,
x = type.beta,
y = native.beta,
style = "equal",
dim = 4)
bivar_map_types_endemic <-
ggplot() +
geom_sf(data = sf_bivar_types_endemic,
aes(geometry = geometry,
fill = bi_class),
color = "white",
size = 0.1,
show.legend = FALSE) +
bi_scale_fill(pal = "DkBlue2",
dim = 4) +
theme_classic()+
theme(
legend.position = "bottom",
legend.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
axis.line = element_blank(),
panel.background = element_rect(fill = NA),
plot.background = element_rect(fill = NA)
)
legend <-
bi_legend(pal = "DkBlue2",
dim = 4,
xlab = "Non endemic",
ylab = "Endemic",
size = )
bivar_map_type_final_endemic <-
ggdraw() +
draw_plot(legend, 0.0, 0.15, 0.25, 0.25) +
draw_plot(bivar_map_types_endemic, 0, 0, 1, 1)Joining all the maps
map_turnover_endemic <-
map_native_endemic_beta+map_type_endemic_beta+bivar_map_type_final_endemic+
patchwork::plot_layout(
design =
"AB
CC"
)+
patchwork::plot_annotation(tag_levels = "a")&
theme(
plot.tag = element_text(face = "bold", hjust = 0, vjust = 1),
plot.tag.position = c(0, 1),
)
map_turnover_endemic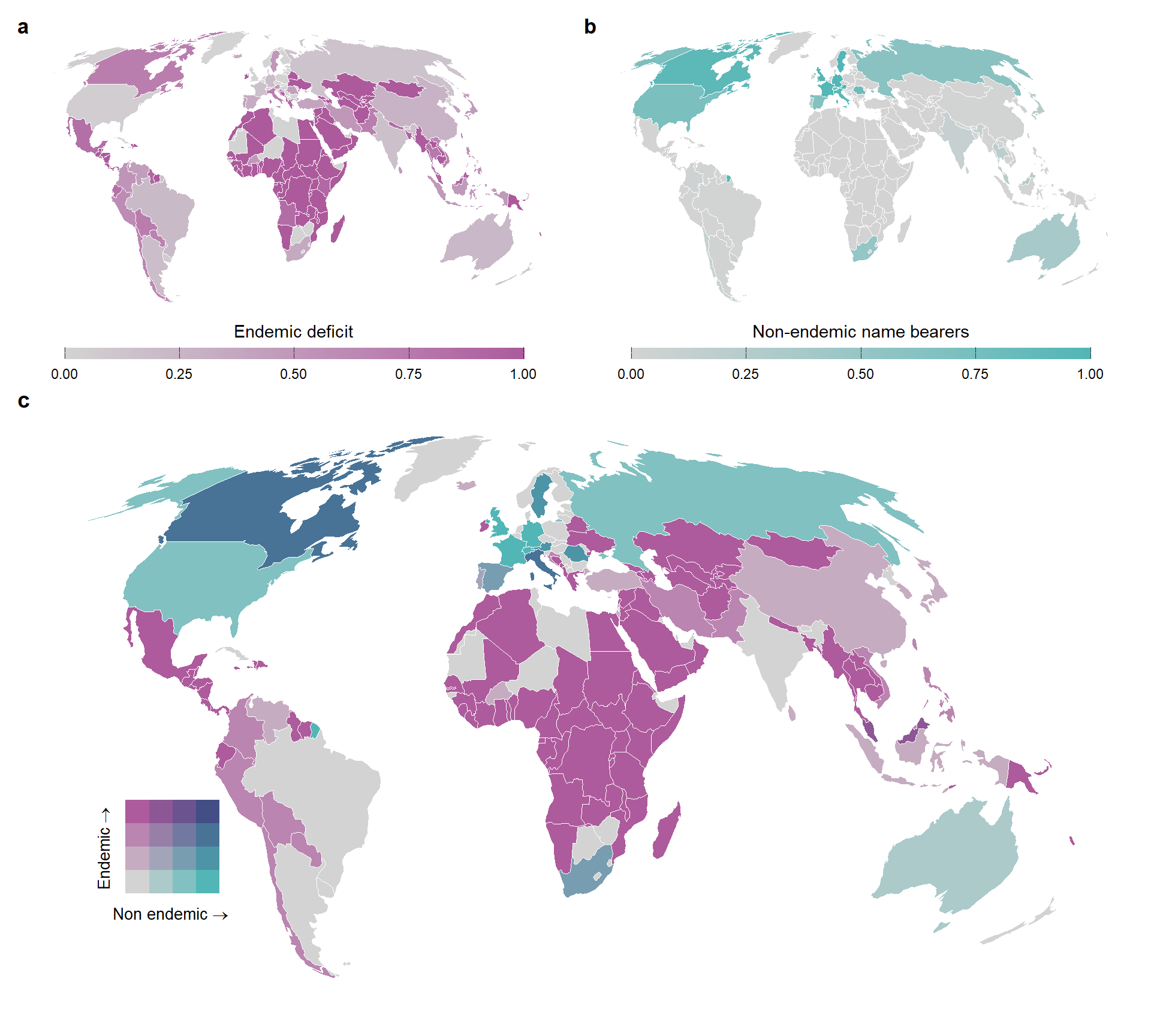
ggsave(here::here("output", "figures", "Fig2_turnover_metrics_endemics.png"),
map_turnover_endemic, dpi=600, width = 10, height = 9)